Sometimes it seems like about half the time I spend in lab is time spent doing mutagenesis (yes, I am my lab's technician, and I like it that way). Quikchange (Now apparently "Quikchange Lighting") is the old standby, but now we have Q5 mutagenesis, and many other options I haven't tried. The drawback to all of these is that they rely on getting a polymerase to amplify an entire plasmid backbone, which is not a simple thing to do. I recently learned (from Chris Campbell in Arshad Desai's lab upstairs) about a way to use Gibson Assembly to make mutagensis, or by extension short insertions or deletions, a whole lot easier by breaking one long PCR into two shorter ones.
When I do mutagenesis, I typically start by using PrimerX to design a pair of complementary primers that should (and sometimes do!) work in a Quikchange-style PCR: 18-24 cycles of PCR, with a long extension time to allow the polymerase to synthesize the entire plasmid backbone. But more often than not, this doesn't work the first, second, or even third time I try it.
A primer pair designed for Quickchange Mutagenesis
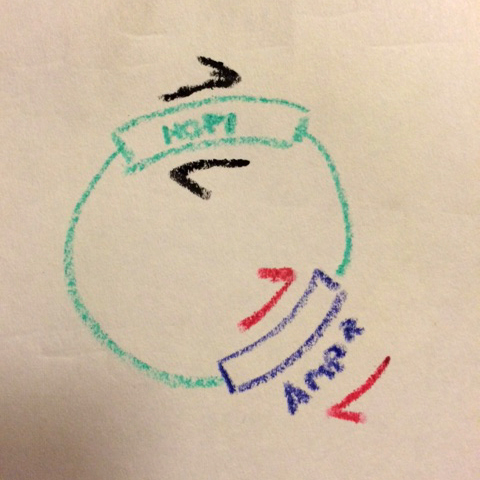
What Chris realized was that almost every plasmid we use has a common feature: the antibiotic resistance gene. Chris designed a pair of complementary primers on the Ampicillin resistance gene in pET3a-type vectors (we also made a pair that prime off the Kanamycin resistance gene in pET28b-type vectors).
Two primer pars now, on opposite sides of the vector...
Then, for every mutation, he runs two reactions with different primer pairs:
(1) The forward primer for the mutation of interest + the reverse primer (#2 below) for the antibiotic resistance gene,
(2) The reverse primer for the mutation of interest + the forward primer (#1 below) for the antibiotic resistance gene.
The PCR products for (1) and (2) are shorter than the whole plasmid, and in my experience so far the reactions are MUCH more reliable (we use Phusion Polymerase almost exclusively). What you are left with is two linear pieces of DNA that overlap by 20-30 bases on each end - exactly what Gibson Assembly is good at sticking together. Gel purify the products from (1) and (2), toss a few nanograms of each in a Gibson Assembly reaction, and transform. Voila, easier mutagenesis.
Boom.
(BTW, I realize that this type of thing becomes obvious once you realize the power of combining Gibson Assembly with creative PCR and primer design - part of the point of this post is to get you thinking about ways to get your own cloning tasks done easier. Intron deletion? Chimeras? Multi-part assembly? Designed-from-scratch biofuel producing superbug? All easier than you think, thanks to Daniel Gibson and Craig Venter. Go San Diego!)
Our primer sequences for Amp and Kan genes, which work anywhere from 45° to 65° annealing temperatures (check the orientation of these on your vector before you use them - our Kan gene is backwards w/r/t our inserts, but the primers for both Amp and Kan are designed so that you use #2 with the mutagenic forward primer, and #1 with the mutagenic reverse primer. Your mileage may vary...):
Isotherm_Amp_primer1 AGAATTATGCAGTGCTGCC
Isotherm_Amp_primer2 GGCAGCACTGCATAATTCT
Isotherm_Kan_primer1 GACAATTACAAACAGGAATCGAATGC
Isotherm_Kan_primer2 GCATTCGATTCCTGTTTGTAATTGTC
Great references for Gibson Assembly:
Gibson, D.G. et.al (2009). Nature Methods. 343-345.
Gibson, D.G. et al. (2010). Nature Methods. 901-903.
Product page and usage guide: New England Biolabs 2X Gibson Assembly Master Mix
Update 10/6/14:
SGI-DNA, a spin-off of Synthetic Genomics here in La Jolla, is now selling a Gibson Assembly kit that they say is much better than "the leading competitor". I haven't tried it, but you can judge for yourself... at the very least, it is less expensive than NEB's kit.